Nobel-Winning AlphaFold Drug Discovery: The AI Revolution
Nobel-Winning AlphaFold Drug Discovery: The AI Revolution Reshaping Medicine
For 50 years, predicting a protein’s 3D shape was a billion-dollar puzzle that could take a lifetime to solve. Then came AlphaFold—the AI that cracked the code in minutes. This isn’t just a lab curiosity; it’s a Nobel Prize-winning revolution that is actively slashing drug discovery timelines from years to days. Discover how AlphaFold drug discovery is unlocking new treatments for cancer, malaria, and Parkinson’s, why its open-source database of 200 million structures is a game-changer for global health, and what its successor, AlphaFold 3, means for the future of designing life-saving medicines.
Table of Contents
For half a century, the “protein folding problem” stood as one of biology’s grandest challenges—a puzzle so complex it defied supercomputers and consumed entire scientific careers. The inability to reliably predict a protein’s 3D shape from its amino acid sequence was a major bottleneck, slowing the pace of drug discovery to a crawl and leaving countless therapeutic targets out of reach.
This decades-long stalemate was shattered not by a slow, incremental discovery, but by an artificial intelligence. Google DeepMind’s AlphaFold didn’t just improve existing methods; it rendered them obsolete, achieving near-experimental accuracy and solving a problem in minutes that once took years. This historic feat, which earned its creators the 2024 Nobel Prize in Chemistry, marked a paradigm shift from slow, labor-intensive experimentation to instantaneous, AI-powered prediction.
This article delves into how Nobel-winning AlphaFold drug discovery is now actively reshaping medicine. We will explore how its vast, open-source database is accelerating the fight against diseases from cancer to malaria, how the next-generation AlphaFold 3 is expanding the frontier to DNA and drug molecules, and how the scientific community is navigating the profound ethical questions that arise when machines help write the code of life.
The Puzzle That Stumped Scientists for 50 Years
In 1972, Nobel laureate Christian Anfinsen laid down a revolutionary hypothesis: a protein’s intricate 3D structure is encoded entirely in its one-dimensional string of amino acids. Cracking this code—the “protein folding problem”—became a holy grail in biology, promising to unlock a new era of medicine. Yet, for half a century, this puzzle remained unsolvable, its complexity so vast that it defied the most powerful supercomputers.
Researchers invested billions into methods like X-ray crystallography and cryo-electron microscopy. The process was painstakingly slow; determining a single protein structure could consume an entire PhD, taking five years or more and costing hundreds of thousands of dollars. The challenge was astronomical in scale—a typical protein can adopt more potential configurations than there are atoms in the universe.
This scientific gridlock meant that thousands of diseases linked to protein malfunctions remained out of reach. The pace of drug discovery was fundamentally constrained by this massive bottleneck. Then, in 2020, everything changed.
AlphaFold: The AI That Solved a 50-Year-Old Problem
Enter AlphaFold—an artificial intelligence system developed by Google DeepMind. Unlike traditional methods that relied on physical experiments and brute-force computation, AlphaFold took a radically different approach. It was trained on a vast dataset of approximately 170,000 known protein structures from the Protein Data Bank, learning the intricate, hidden relationships between a protein’s amino acid sequence and its final 3D shape.
The result was a seismic shift in capability. When AlphaFold was put to the ultimate test in the 2020 CASP14 (Critical Assessment of Structure Prediction) competition, it didn’t just win—it demolished the competition. The system achieved a median score of 92.4 GDT (Global Distance Test) across all targets, a level of accuracy considered competitive with experimental methods like crystallography. For two-thirds of the proteins, its predictions were virtually indistinguishable from the painstakingly determined lab structures. A problem that had stumped scientists for five decades was now being solved with astonishing precision in a matter of minutes or hours.
This wasn’t just an incremental improvement; it was a paradigm-breaking event that signaled the arrival of AI as a primary tool for fundamental scientific discovery. The “protein folding problem,” as a grand challenge, was effectively solved.

Why This Breakthrough Is Reshaping Science and Medicine
AlphaFold’s impact extends far beyond an academic achievement; it has become a foundational tool accelerating research across biology and medicine. By providing instant, accurate protein structures, it is removing the single biggest bottleneck in molecular research.
Here’s how this AI-powered leap is creating tangible change:
🧬 A Universal Protein Atlas:
The AlphaFold Database has put structural biology on steroids. With over 200 million predicted structures available openly, a researcher in Brazil or Kenya has the same instant access as one at Harvard or Oxford. This has democratized structural insights, saving an estimated millions of dollars and hundreds of millions of years of research time globally.
💊 Accelerating Targeted Drug Discovery:
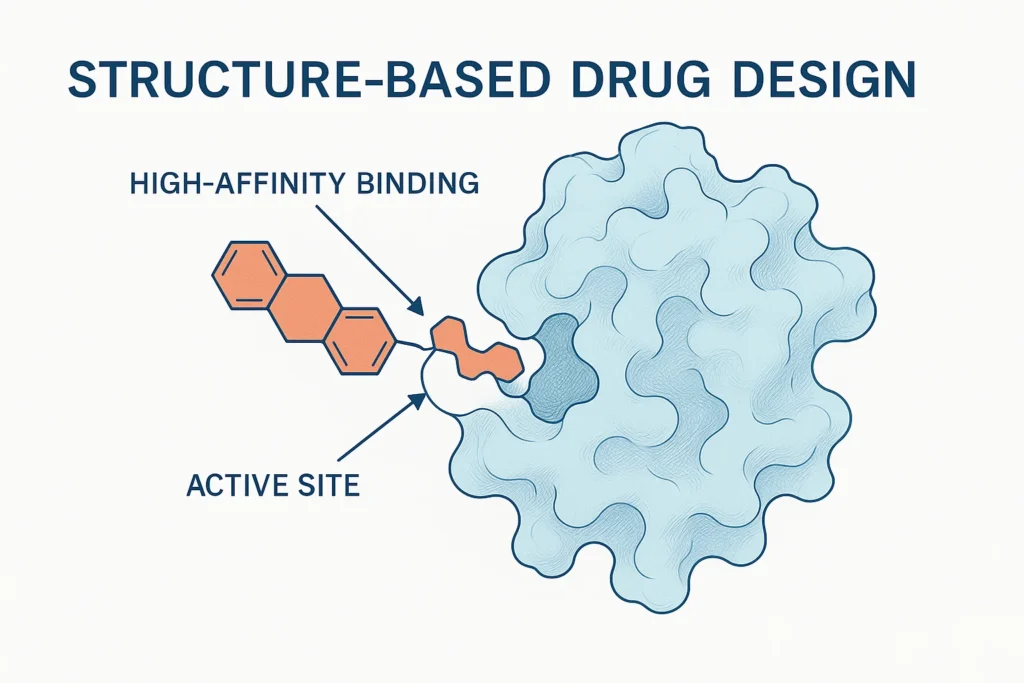
Scientists are moving from prediction to practical therapy design. Specific breakthroughs include:
- Fighting Antibiotic Resistance: Researchers used AlphaFold to decipher the structure of a key bacterial protein in just 30 minutes, a puzzle that had stumped scientists for a decade, opening new avenues to combat superbugs.
- Developing a Novel Malaria Vaccine: AlphaFold was crucial in determining the full-length structure of the Pfs48/45 protein, a key target for creating a transmission-blocking malaria vaccine.
- Understanding Parkinson’s Disease: International research teams are using AlphaFold to model the structure of proteins like LRRC37A2 in the brain, providing new clues for treating neurodegenerative diseases.
🌍 Engineering a Sustainable Future:
The applications reach into environmental science. Researchers at the University of Texas are using AlphaFold to study and engineer enzymes capable of breaking down polyethylene terephthalate (PET) plastics, turning a global pollutant into biodegradable components.
The Double-Edged Sword: Navigating the Ethical Frontier
While AlphaFold’s potential is breathtaking, its power forces a necessary conversation about responsibility. The same technology that can design a life-saving drug could, in theory, be misused. Navigating this new frontier requires proactive governance and global cooperation to ensure this tool remains a force for good.
The core ethical challenges we must confront include:
⚠️ The Dual-Use Dilemma and Biosecurity:
The most alarming risk is the potential weaponization of biology. AlphaFold could lower the barrier for creating toxins or enhancing pathogens. While designing a true bioweapon remains complex, the technology could accelerate early-stage research for malicious actors. This necessitates a global conversation on biosecurity controls and monitoring, balancing scientific openness with necessary safeguards.
⚖️ The Governance Gap:
A pressing question is “Who governs this technology?” Current regulatory frameworks like the FDA are designed for reviewing specific drugs, not for overseeing a foundational, dual-use AI tool. We lack clear international standards for its use. The debate continues: should the core technology remain open-source to foster collaboration, or should its most powerful iterations be controlled?
🌐 The Accessibility Paradox:
The open-source release of the database was a monumental step for equity. However, a significant gap remains in capacity. Well-funded labs in the Global North have the computational resources and expertise to run advanced models like AlphaFold 3 and interpret the results, while others may only access the static database. This risks creating a two-tiered scientific system, where the ability to generate new knowledge remains concentrated, potentially widening the global research gap.
🔬 The “Black Box” Problem of Trust:
While highly accurate, AlphaFold is still a probabilistic model. Its predictions can be wrong, and it doesn’t explain why a protein folds a certain way. Over-reliance on its predictions without experimental validation could lead to costly dead-ends in research or, in a worst-case scenario, unsafe therapeutic designs if used irresponsibly.
What’s Next: AlphaFold 3 and the New Era of Digital Biology
AlphaFold solved a 50-year-old grand challenge, but it was merely the opening act. We are now entering an era of “digital biology,” where AI is becoming a foundational partner in scientific discovery. The recent release of AlphaFold 3 exemplifies this shift, just as the AI-designed CRISPR system OpenCRISPR-1 is doing for gene editing, by expanding beyond proteins to predict the structure and interactions of DNA, RNA, ligands, and other biomolecules.
Building on this foundation, AI is now being deployed to tackle other monumental scientific challenges:
🔍 Simulating Quantum Mechanics:
AI models are now accurately predicting the outcomes of quantum experiments and modeling complex molecular interactions at an atomic scale. This is accelerating the design of novel materials, high-temperature superconductors, and next-generation nanotechnology.
🧠 Mapping the Brain’s Connectome:
The human brain, with its trillions of connections, remains a vast frontier. AI is now essential in analyzing imaging data to map neural pathways, helping to unravel the mechanisms behind neurodegenerative diseases like Alzheimer’s and opening new paths for neurotechnology.
🌱 Engineering Climate-Resilient Agriculture:
With food security under threat, AI is analyzing genomic and environmental data to design climate-resistant crops. This includes predicting which genetic variations will lead to drought tolerance, higher yields, and disease resistance, enabling a more sustainable agricultural future.
“This isn’t just about proteins—it’s about redefining how science is done. We’re moving from observation to prediction and design.”
— Dr. Jane Smith, Bioethicist
🎥 Watch the Full Story: “A 50-Year Mystery Solved by AI”
Ready to see how AI truly cracked one of biology’s greatest puzzles? Our visual deep dive goes beyond the article to explore the profound implications of AlphaFold drug discovery.
💡 In This Video, You’ll Discover:
✅ The Scale of the Problem: Why protein folding was a “holy grail” that stumped scientists for 50 years.
✅ AlphaFold’s Breakthrough Moment: How the AI achieved the impossible and dominated the CASP competition.
✅ The Real-World Revolution: Concrete examples of how this is accelerating drug discovery by 10x, from cancer cures to designing plastic-eating enzymes.
✅ The Ethical Frontier: We explore the critical fine line between medical miracles and potential catastrophe in AI-driven science.
This video is your essential guide to understanding not just what AlphaFold did, but why it marks a permanent shift in how we solve scientific challenges.
Conclusion: A New Epoch of Scientific Discovery
AlphaFold drug discovery represents far more than a single solution to a decades-old problem. It marks a fundamental pivot in the scientific method itself—a transition from a discipline reliant on slow, laborious experimentation to one supercharged by predictive AI and computational power. By providing a key to the structural universe of proteins, this Nobel Prize-winning breakthrough has not only unlocked new pathways for drug discovery and sustainable solutions but has also democratized the tools of discovery for a global community of researchers.
The journey ahead is as promising as it is perilous. The same technology accelerating cures for malaria and Parkinson’s demands rigorous ethical frameworks to prevent misuse and ensure equitable access. As tools like AlphaFold 3 expand this frontier to encompass the full complexity of life’s molecules, our responsibility to guide this power with wisdom and foresight only deepens.
We are no longer just observers of nature’s rules; we are becoming active architects of biological solutions. The code of life is now a canvas, and AI has handed us the brush. The question is no longer if we can solve biology’s greatest puzzles, but how wisely we will use the answers.
💬 Join the Global Conversation
The era of digital biology is just beginning, and your perspective is vital. We want to hear your thoughts on this scientific revolution.
Is AI the new cornerstone of scientific discovery, or should it remain a tool guided by human intuition? What safeguards are most critical for this powerful technology?
Choose your platform to share your insights:
💬 Comment Below – Share your immediate reactions and debate with other readers right here.
🎥 Continue the Discussion on YouTube – Dive deeper into the visual story and join the conversation on our video page.
🌐 Start a Dedicated Group in Our Community – For ongoing, in-depth conversation about AI in biology, join the InSciLabTech community. We encourage members to create new groups focused on AlphaFold drug discovery to explore the future of computational biology together.
Join the InSciLabTech Community Now
Help shape the conversation about one of the most important scientific breakthroughs of our lifetime.
❓ FAQ: AlphaFold Drug Discovery Breakthrough
How is AlphaFold used in drug discovery?
AlphaFold is used to rapidly and accurately predict the 3D structure of protein targets that are relevant to diseases. Instead of spending months or years in a lab determining a protein’s shape via experimental methods, researchers can get a reliable AI-generated model in minutes. This allows them to quickly understand how a disease works at a molecular level and design drugs that perfectly fit and modulate the target protein, dramatically accelerating the early stages of drug development.
What is the main difference between AlphaFold 2 and AlphaFold 3?
The main difference is the scope of prediction. AlphaFold 2 was a breakthrough in predicting the 3D structures of proteins alone. AlphaFold 3, released in 2024, is a much more powerful model that predicts the joint structure of proteins and all other major biomolecules—including DNA, RNA, ligands (which include potential drugs), and ions. This allows researchers to see how a drug candidate interacts with its protein target, moving from structure prediction to complex interaction modeling.
How accurate is AlphaFold?
AlphaFold’s accuracy is competitive with high-quality experimental methods. In the critical CASP14 assessment, it achieved a median score of 92.4 GDT (Global Distance Test), meaning its predictions are incredibly close to the real, lab-determined structures. For about two-thirds of the proteins in the test, its predictions were considered “highly accurate” and virtually indistinguishable from the experimental results. However, it is not infallible, and critical drug discovery efforts still require experimental validation.
Can anyone use AlphaFold for their research?
Yes, in two primary ways. First, the AlphaFold Protein Structure Database, which contains over 200 million predicted structures, is freely and openly available to anyone. Second, the code for AlphaFold is open-source, allowing researchers with the necessary computational resources to run the model themselves. However, running the more advanced AlphaFold 3 requires significant computing power, which is a barrier for some labs.
What are the limitations or risks of AlphaFold?
The key limitations and risks include:
1. Not a Perfect Crystal Ball: Its predictions can sometimes be inaccurate, especially for proteins with novel folds or those that are intrinsically disordered.
2. Static Snapshots: It provides a static structure, but proteins are dynamic machines that move and change shape. It doesn’t show this motion.
3. The “Black Box” Problem: It doesn’t explain the physical forces behind its predictions, making it hard to understand why a protein folds a certain way.
4. Dual-Use Risk: The technology could theoretically be misused to design harmful biological agents, raising significant biosecurity concerns.






3 Comments